This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.
Targeted Proteomics
at the Institute for Systems Biology
Targeted Proteomics
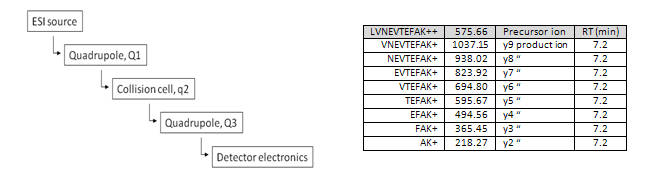
Targeted proteomics experiments are hypothesis driven efforts to quantitate the relative amounts of protein in a sample. In these experiments, the proteins are already known and no discovery is needed. A database link is consulted to determine the relevant proteotypic peptides and fragmentation patterns for the proteins of interest. This information is used to construct a transition list for multiple selected reaction monitoring analyses employing triple quadrupoles. Quantitation is performed by comparing the native peptide abundance to the abundance of an isotopically enriched peptide spiked into the sample during preparation. There are five triple quadrupoles in the ISB program.
SRM and MRM
Selected Reaction Monitoring, sometimes called Multiple Reaction Monitoring, is a high throughput mass spectrometry technique for quantitative analysis. It requires a triple quadrupole and knowledge about the ions generated when a peptide is ionized and fragmented. The first ion, the precursor, is created in the ESI source as it enters the mass spectrometer. It is selected by the first quadrupole (Q1) and delivered to the collision cell (q2) where it is fragmented into its smaller, product ions. These are selected one at a time by the third quadrupole (Q3) and delivered individually to the detector where they are counted.
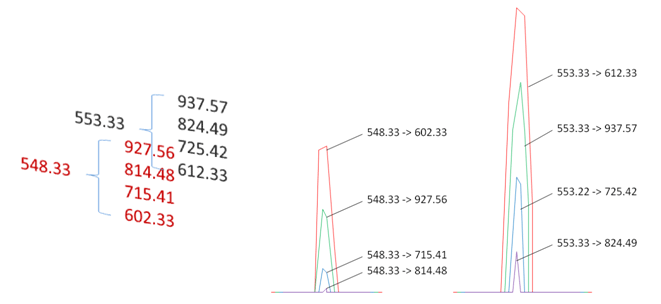
To quantitate using SRM, isotopically labeled versions of the peptides, termed “heavy” peptides, are added to the sample as a standard addition. The native peptides or “light” peptides and are produced by digesting the proteins isolated from the sample. The heavy peptides are artificial and do not exist in nature. However, they are chromatographically identical to the light peptides and elute at the same retention time. Thus, for every eluting peptide there is intensity information for multiple precursor->product ion transitions, including the set for the heavy peptide whose concentration is known. By calculating the ratio between the light and the heavy peptide transitions, the concentration of the native peptide is determined. This is taken to be a relative measure of the protein in the initial sample.
Depending on the chromatographic conditions, it is not uncommon to have several thousand transitions (precursor-product ion pairs) belonging to hundreds of peptides. The triple quadrupole multiplexes the transition list and analyzes each precursor and fragment-ion transition in turn. It does this very rapidly and hundreds of peptides can be analyzed in a single run. In a 24 hour period, analytical data for thousand peptides can be collected. No computer search is necessary and the data can be rapidly analyzed by a number of quantitative software packages.